Data Preparation
While R itself has some powerful tools for data preparation, dplyr package makes such tasks much efficient and easier.
We will use cake dataset - you can download it from here. First, import dplyr.
> library(dplyr)
Filtering Data
Selecting subsets or slices from dataframes is achieved through the filter function of dplyr. Let's say we want to select part of the dataframe where only vanilla flavor is listed.
> filter(cake, Flavor == 'Vanilla')
# A tibble: 6 x 6 LastName Age PresentScore TasteScore Flavor Layers 1 Orlando 27 93 80 Vanilla 1 2 Goldston 46 68 75 Vanilla 1 3 Roe 38 79 73 Vanilla 2 4 Nguyen 57 77 84 Vanilla NA 5 Byron 62 72 87 Vanilla 2 6 Conrad 69 85 94 Vanilla 1
We can also put more than one conditions.
> filter(cake, Flavor == 'Vanilla', Layers == '1')
# A tibble: 3 x 6 LastName Age PresentScore TasteScore Flavor Layers 1 Orlando 27 93 80 Vanilla 1 2 Goldston 46 68 75 Vanilla 1 3 Conrad 69 85 94 Vanilla 1
Above was an example of 'AND' operator. We could have achieved the same result with the following:
> filter(cake, Flavor == 'Vanilla' & Layers == '1')
Similarly we can use an 'OR' operator with '|':
> filter(cake, Flavor == 'Vanilla' | Layers == '1')
# A tibble: 12 x 6 LastName Age PresentScore TasteScore Flavor Layers 1 Orlando 27 93 80 Vanilla 1 2 Goldston 46 68 75 Vanilla 1 3 Roe 38 79 73 Vanilla 2 4 Strickland 19 82 79 Chocolate 1 5 Nguyen 57 77 84 Vanilla NA 6 Hildenbrand 33 81 83 Chocolate 1 7 Byron 62 72 87 Vanilla 2 8 Sanders 26 56 79 Chocolate 1 9 Jaeger 43 66 74 "" 1 10 Conrad 69 85 94 Vanilla 1 11 Anderson 27 87 85 Chocolate 1 12 Merritt 62 73 84 Chocolate 1
Following example will show selecting 'NA' values (as we cannot simply use '==NA')
> filter(cake, is.na(Layers) == TRUE)
# A tibble: 2 x 6 LastName Age PresentScore TasteScore Flavor Layers 1 Larsen 23 77 84 Chocolate NA 2 Nguyen 57 77 84 Vanilla NA
Sorting Rows
Sorting rows is done via arrange function of dplyr.
> arrange(cake, LastName)
# A tibble: 20 x 6 LastName Age PresentScore TasteScore Flavor Layers 1 Anderson 27 87 85 Chocolate 1 2 Becker 36 62 83 Spice 2 3 Byron 62 72 87 Vanilla 2 4 Conrad 69 85 94 Vanilla 1 5 Davis 51 86 91 Spice 3 6 Davis 28 69 75 Chocolate 2 7 Goldston 46 68 75 Vanilla 1 8 Hildenbrand 33 81 83 Chocolate 1 9 Jaeger 43 66 74 "" 1 10 Larsen 23 77 84 Chocolate NA 11 Matthew 42 81 92 Chocolate 2 12 Merritt 62 73 84 Chocolate 1 13 Nguyen 57 77 84 Vanilla NA 14 Orlando 27 93 80 Vanilla 1 15 Ramey 32 84 72 Rum 2 16 Roe 38 79 73 Vanilla 2 17 Rossburger 28 78 81 Spice 2 18 Sanders 26 56 79 Chocolate 1 19 Strickland 19 82 79 Chocolate 1 20 Walters 55 67 72 Chocolate 2
Default sorting style is ascending. To change it:
> arrange(cake, desc(LastName))
# A tibble: 20 x 6 LastName Age PresentScore TasteScore Flavor Layers 1 Walters 55 67 72 Chocolate 2 2 Strickland 19 82 79 Chocolate 1 3 Sanders 26 56 79 Chocolate 1 4 Rossburger 28 78 81 Spice 2 5 Roe 38 79 73 Vanilla 2 6 Ramey 32 84 72 Rum 2 7 Orlando 27 93 80 Vanilla 1 8 Nguyen 57 77 84 Vanilla NA 9 Merritt 62 73 84 Chocolate 1 10 Matthew 42 81 92 Chocolate 2 11 Larsen 23 77 84 Chocolate NA 12 Jaeger 43 66 74 "" 1 13 Hildenbrand 33 81 83 Chocolate 1 14 Goldston 46 68 75 Vanilla 1 15 Davis 51 86 91 Spice 3 16 Davis 28 69 75 Chocolate 2 17 Conrad 69 85 94 Vanilla 1 18 Byron 62 72 87 Vanilla 2 19 Becker 36 62 83 Spice 2 20 Anderson 27 87 85 Chocolate 1
Selecting Subsets
There will be times where we are interested in certain columns but not the others. select function of dplyr helps us to achieve just that. Say we would like to get a new subset from LastName and PresentScore. Following will achieve that:
> select(cake, LastName, PresentScore)
# A tibble: 20 x 2 LastName PresentScore 1 Orlando 93 2 Ramey 84 3 Goldston 68 4 Roe 79 5 Larsen 77 6 Davis 86 7 Strickland 82 8 Nguyen 77 9 Hildenbrand 81 10 Byron 72 11 Sanders 56 12 Jaeger 66 13 Davis 69 14 Conrad 85 15 Walters 67 16 Rossburger 78 17 Matthew 81 18 Becker 62 19 Anderson 87 20 Merritt 73
To select multiple columns we use colon(:) operator (similar to -- in SAS).
> select(cake, LastName:PresentScore)
# A tibble: 20 x 3 LastName Age PresentScore 1 Orlando 27 93 2 Ramey 32 84 3 Goldston 46 68 4 Roe 38 79 5 Larsen 23 77 6 Davis 51 86 7 Strickland 19 82 8 Nguyen 57 77 9 Hildenbrand 33 81 10 Byron 62 72 11 Sanders 26 56 12 Jaeger 43 66 13 Davis 28 69 14 Conrad 69 85 15 Walters 55 67 16 Rossburger 28 78 17 Matthew 42 81 18 Becker 36 62 19 Anderson 27 87 20 Merritt 62 73
Renaming Columns
Renaming columns is done with rename function of dplyr.
> rename(cake,PScore = PresentScore)
Note that the order is important: new_name = old_name will work, old_name = new_name won't.
# A tibble: 20 x 6 LastName Age PScore TasteScore Flavor Layers 1 Orlando 27 93 80 Vanilla 1 2 Ramey 32 84 72 Rum 2 3 Goldston 46 68 75 Vanilla 1 4 Roe 38 79 73 Vanilla 2 5 Larsen 23 77 84 Chocolate NA 6 Davis 51 86 91 Spice 3 7 Strickland 19 82 79 Chocolate 1 8 Nguyen 57 77 84 Vanilla NA 9 Hildenbrand 33 81 83 Chocolate 1 10 Byron 62 72 87 Vanilla 2 11 Sanders 26 56 79 Chocolate 1 12 Jaeger 43 66 74 "" 1 13 Davis 28 69 75 Chocolate 2 14 Conrad 69 85 94 Vanilla 1 15 Walters 55 67 72 Chocolate 2 16 Rossburger 28 78 81 Spice 2 17 Matthew 42 81 92 Chocolate 2 18 Becker 36 62 83 Spice 2 19 Anderson 27 87 85 Chocolate 1 20 Merritt 62 73 84 Chocolate 1
Adding Columns
We can add new columns with mutate function.
> mutate(cake, AvgScore = (PresentScore + TasteScore) / 2)
# A tibble: 20 x 7 LastName Age PresentScore TasteScore Flavor Layers AvgScore 1 Orlando 27 93 80 Vanilla 1 86.5 2 Ramey 32 84 72 Rum 2 78 3 Goldston 46 68 75 Vanilla 1 71.5 4 Roe 38 79 73 Vanilla 2 76 5 Larsen 23 77 84 Chocolate NA 80.5 6 Davis 51 86 91 Spice 3 88.5 7 Strickland 19 82 79 Chocolate 1 80.5 8 Nguyen 57 77 84 Vanilla NA 80.5 9 Hildenbrand 33 81 83 Chocolate 1 82 10 Byron 62 72 87 Vanilla 2 79.5 11 Sanders 26 56 79 Chocolate 1 67.5 12 Jaeger 43 66 74 "" 1 70 13 Davis 28 69 75 Chocolate 2 72 14 Conrad 69 85 94 Vanilla 1 89.5 15 Walters 55 67 72 Chocolate 2 69.5 16 Rossburger 28 78 81 Spice 2 79.5 17 Matthew 42 81 92 Chocolate 2 86.5 18 Becker 36 62 83 Spice 2 72.5 19 Anderson 27 87 85 Chocolate 1 86 20 Merritt 62 73 84 Chocolate 1 78.5
Summarizing Data
Oftentimes we would like to have a summary of our dataset before venturing into further statistical analysis. summarize and group_by functions group and summarize data frames.
> summarize(group_by(cake, Flavor), mean(PresentScore, na.rm=TRUE))
# A tibble: 5 x 2 Flavor `mean(PresentScore, na.rm = TRUE)` 1 "" 66 2 Chocolate 74.8 3 Rum 84 4 Spice 75.3 5 Vanilla 79
Pipelines
Pipelines help us to input multiple commands into single one. Pipeline operator in R is %>%. For example we would like to get averages of different flavors excluding empty flavors and NA layers:
> filter(cake, Flavor != '', is.na(Layers) == FALSE) %>%
+ group_by(Flavor) %>%
+ summarize(avgscore = mean(PresentScore))
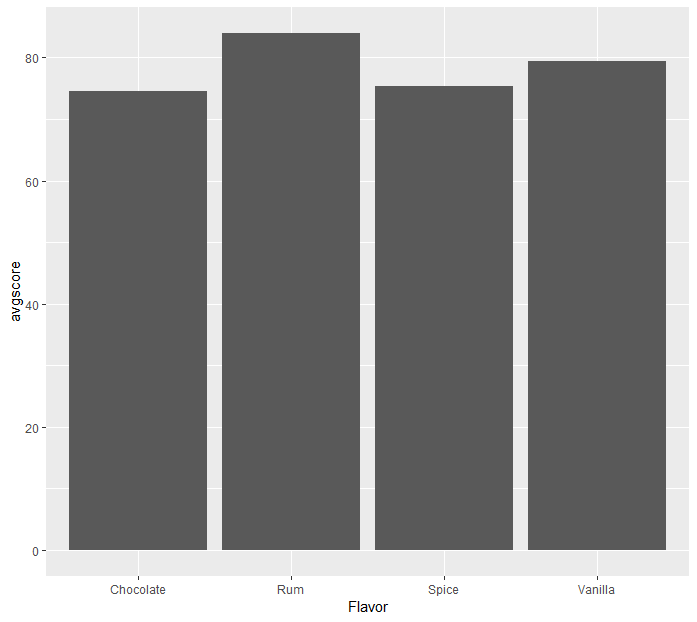
# A tibble: 4 x 2 Flavor avgscore 1 Chocolate 74.5 2 Rum 84 3 Spice 75.3 4 Vanilla 79.4
We can even further extend our pipeline to ggplot:
> filter(cake, Flavor != '', is.na(Layers) == FALSE) %>%
+ group_by(Flavor) %>%
+ summarize(avgscore = mean(PresentScore)) %>%
+ ggplot(mapping = aes(x=Flavor, y=avgscore)) + geom_bar(stat='identity')
Transposing Data
reshape2 is one of the best packages around to transform our data. Here is an example: suppose we have a dataframe as below.
| Bag | Color | n | nn |
|---|---|---|---|
| A | blue | 8 | 16 |
| A | green | 14 | 28 |
| A | red | 8 | 16 |
| B | blue | 16 | 32 |
| B | green | 7 | 14 |
| B | red | 7 | 14 |
To transform this dataframe into the following,
| Bag | Color | variable | value |
|---|---|---|---|
| A | blue | n | 8 |
| A | green | n | 14 |
| A | red | n | 8 |
| B | blue | n | 16 |
| B | green | n | 7 |
| B | red | n | 7 |
| A | blue | nn | 16 |
| A | green | nn | 28 |
| A | red | nn | 16 |
| B | blue | nn | 32 |
| B | green | nn | 14 |
| B | red | nn | 14 |
we will use the melt function of reshape2 package:
> library(reshape2)
> new <- melt(old, id=c('Bag','Color'), na.rm=TRUE)
Any variable that is specified in 'id' argument will stay where they are, rest of the columns will be packed into two columns.
To transform this data back to its original form:
> old <- dcast(new, Bag+Color~variable, value.var = 'value')
Similarly this statement says 'keep Bag and Color in place, distribute values in variable as columns and fill the values with the value of n'.
Splitting Columns
Sometimes we come across columns packing more than a single variable. For example, take a look at table 'bags' below:
| Bag | n | nn |
|---|---|---|
| A Blue | 8 | 64 |
| A Green | 14 | 196 |
| A Red | 8 | 64 |
| B Blue | 16 | 256 |
| B Green | 7 | 49 |
| B Red | 7 | 49 |
Column 'Bag' includes both the type and color of bags. We need to separate them into two columns instead. We will use reshape2 package again for this purpose.
> library(reshape2)
> colsplit(bags$Bag, ' ', c('Type', 'Color')) %>%
+ data.frame(bags) %>%
+ select(Type, Color, n, nn)
Type Color n nn 1 A blue 8 64 2 A green 14 196 3 A red 8 64 4 B blue 16 256 5 B green 7 49 6 B red 7 49
Here is how the above code works: colsplit function separates data in 'Bag' column (data which is separated by space) into a matrix with two columns, 'Type' and 'Color'. This generated matrix is then pipelined to data.frame function which combines it with the original dataframe and output it as a new dataframe. This new dataframe is subset by the tidyr's select function into the final dataframe.
Export Tables
write.csv is the function of R to export any table in csv format. Here's how we do it:
> write.csv(mytable, 'D:\\teaching\\mytable.csv')
Note that the default options save row names. If you don't want to save row names, you need to specify it:
> write.csv(mytable, 'D:\\teaching\\mytable.csv', row.names=FALSE)
Missing values by default are saved as 'NA'. If you want to export just a blank cell instead:
> write.csv(mytable, 'D:\\teaching\\mytable.csv', row.names=FALSE, na="")







Leave a Comment